Numpy And Scipy笔记(三)
文章目录
|
|
SciKit
本章主要讨论:
Scikitimage:比`scipy.ndimage`更强的图像处理模块。
Scikitlearn:机器学习包。
Scikit-Image
SciPy和ndimage类包含了非常多的处理多维数据的工具,如过滤(高斯平滑)傅利叶变换,形态(二进制侵蚀),插值和测量。可以组合这些工具进行更复杂的处理。Scikit-image正是这一类高级模块,它包含了颜色空间转换、图像强度调整算法、特征检测、锐化和降噪,图像读写等。
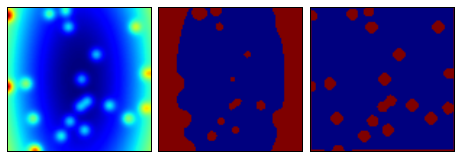
动态阀值
图像处理中的常见的问题是对图像内容进行分块。通常的阀值技术在图像背景色较为平坦时工作得较好。但实际上图像的背景经常是变化的。这种情况下使用scikit-image提供的自适应阀值就能比较容易处理。
下例我们使用生成的图像,这个图像的背景不是统一的,图像上包含随机放置的点。使用自适应阀值技术我们可以从背景中提取出这些白点。
|
|
/Users/zhujie/.virtualenvs/deeplearn/lib/python2.7/site-packages/skimage/filter/__init__.py:6: skimage_deprecation: The `skimage.filter` module has been renamed to `skimage.filters`. This placeholder module will be removed in v0.13.
warn(skimage_deprecation('The `skimage.filter` module has been renamed '
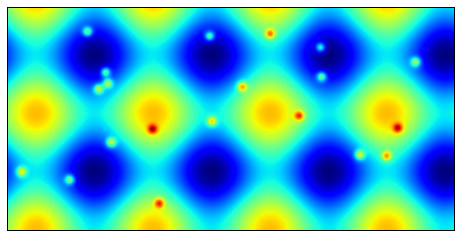
局部极大值
通过使用skiimage.morphology.is_local_maximum可以查找出局部极值。可以用于在图像中查找最亮或最暗的点。
|
|
Scikit-Learn
线性回归
Scikit-learn包含了大量工具,如Lasso和岭回归。这里只演示普通最小二乘回归函数。
$$
\min_{w} \lVert X \beta - y \rVert
$$
$w$ 是系数集,它的数量取决于数据的维度,$ N(系数) = MD - 1,M > 1$ 并且为整数。下例计算3D空间中的一个线性拟合平面,使用LinearRegression来训练数据集。
|
|
[-10.25691752 90.5463984 ]
[ 350.86036386]
0.949827492261
/Users/zhujie/.virtualenvs/deeplearn/lib/python2.7/site-packages/sklearn/utils/validation.py:386: DeprecationWarning: Passing 1d arrays as data is deprecated in 0.17 and willraise ValueError in 0.19. Reshape your data either using X.reshape(-1, 1) if your data has a single feature or X.reshape(1, -1) if it contains a single sample.
DeprecationWarning)
<IPython.core.display.Javascript object>
聚类
SciPy提供了两个用于聚类分析的包矢量量化(kmeans)和分层聚类(hierarchy)。Scikit-learn与SciPy中的kmeans类似功能的是DBSCAN算法,它用于通过给定半径查找核心点,它比kmeans相比,对包含噪声的数据处理效果更好。
|
|
文章作者 Jamsa
上次更新 2017-03-28